RESEARCH
The Gregory lab is involved in several lab-based and collaborative research projects that focus on identifying the genomic, genetic, and epigenetic underpinnings of complex neurological diseases.
Brain tumors:
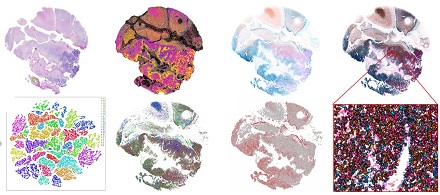
As early adopters of single cell and spatial expression profiling approaches, the Gregory lab and the Molecular Genomics Core is applying early access in situ sequencing technologies to identify the signatures of brain tumor development and progression. These technologies have the capacity to profile RNA expression at true single cell level resolution and in doing so unravel the heterogeneity of the tumor microenvironment using panels of canonical markers of cell type/state to elucidate the cell-cell and cell-neighborhood interaction analyses.
We are combining single nuclei/cell modalities and spatial RNA/protein approaches with traditional whole genome and exome sequencing, epigenetic profiling, and bulk RNA-Seq transcriptome methods to underpin the next phase of brain tumor research. Together with basic and clinical scientists across Duke, the Gregory lab is a founding member of the new Brain Tumor Omics Program developed in conjunction with Dr. David Ashley, Director of the Preston Robert Tisch Brain Tumor Center.
Multiple Sclerosis:
Dr. Gregory is Principal Investigator of the MURDOCK_MS study which includes ~1,000 multiple sclerosis patients. This cohort has been used to identify multi-omic biomarkers to facilitate reclassification of the disease (Cote et al 2019, Multiple Sclerosis and Related Disorders), and to assess MS progression using ultra-high sensitivity protein assays in a cohort of primary progressive MS patients (Profiling serum neurofilament light chain and glial fibrillary acidic protein in primary progressive multiple sclerosis).
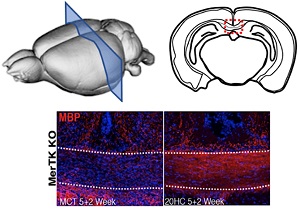
The Gregory lab is also exploring the efficacy of a novel hydroxyl-cholesterol (HC) treatment of remyelination in MS. Together with Drs. Eric Benner (Duke, Pediatrics), Mari Shinohara (Duke, Immunology), and Glenn Matsushima (UNC-CH, Microbiology and Immunology), this transformative study is trying to understand the cellular mechanisms in which HCs trigger the differentiation of neural progenitor cells into oligodendrocyte progenitor cells (OPCs) and/or OPCs into oligodendrocytes, the cell that is critical for triggering remyelination in MS. Early data provides exciting evidence of remyelination in the context of a demyelinating pre-clinical model of MS.
Autism:
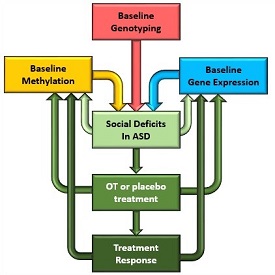
Recent CDC estimates suggest that autism affects more than one in 68 children in the US. The Gregory lab is using genetic and epigenetic approaches to understand the mechanisms driving core deficits associated with autism at developmental and therapeutic levels via oxytocin treatment in pre-clinical and human trials with Dr. Lin Sikich. In collaboration with Dr. Sheryl Moy (UNC, Psychiatry) and Dr. Yong-Hui Jiang (Yale, Medical genetics), Dr. Gregory's lab is exploring the mechanisms of oxytocin response in a clinical trial of autistic children, to refine the epigenetic regulation of the oxytocin receptor (OXTR), and to understand the molecular underpinnings of oxytocin effects and societal interaction in an animal model of autism. The findings will provide valuable insight into the mode of action of oxytocin response in specific regions of the brain that will be applicable to clinical trials of oxytocin treatment in numerous psychosocial phenotypes, including autism.
Alzheimer’s Disease:
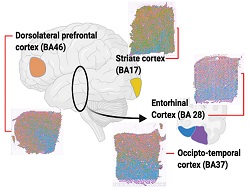
The Gregory lab, together with collaborators Drs. Jerry Wang (Department of Pathology) and Dianne Cruz (Department of Psychiatry), is using spatial transcriptomics to characterize the RNA expression changes that occur with the progression of Alzheimer’s disease. These data are correlated with the microarchitecture of the brain and disease pathology to understand the molecular changes associated with the disease. We are also pairing these data with a long-read sequence platform to explore RNA splicing in a spatial context to determine the effects of AD pathology in isoform production.
SUMMARY
Dr. Gregory is a tenured Professor and Director of the Brain Tumor Omics Program (BTOP) in the Duke Department of Neurosurgery, the Vice Chair of Research in the Department of Neurology, and Director of the Molecular Genomics Core at the Duke Molecular Physiology Institute (DMPI). As a neurogenomicist, Dr. Gregory applies his experience gained from leading the sequencing of chromosome 1 for the Human Genome Project to elucidating the mechanisms underlying multi-factorial diseases using genetic, genomic, and epigenetic approaches. Dr. Gregory’s primary areas of research involve understanding the molecular processes associated with disease development and progression in brain tumors and Alzheimer’s disease, novel drug induced white matter injury repair in multiple sclerosis, and social and behavioral response to oxytocin treatment animal models of autism. He is broadly regarded across Duke as a leader in the development of novel single cell and spatial molecular technologies towards understanding the pathogenic mechanisms of disease development. Dr. Gregory is also the Section Chair of Genomics and Epigenetics at the DMPI and Director of the Duke Center of Autoimmunity and MS in the Department of Neurology.
BSc, Royal Melbourne Institute of Technology, Melbourne, Australia
PhD, Wellcome Trust Sanger Institute, Hinxton, United Kingdom
PUBLICATIONS
LINKS
News Articles
DMPI Faculty Appointments and Promotions
Simon Gregory among group of international members selected to 10x Genomics CTRN
Duke Single Cell Initiative Seminar Series
LAB NEWS
- Simon moderated a panel on gene-editing human embryos following the uproar following the claim of Dr. He Jiankui that he made the world's first genetically edited twins.
- Simon is quoted in an important new study regarding autism risks dependent on the age of the parents.
LAB MEMBERS

Originally from Massachusetts, Karen has been at Duke since 1999 and is the Assistant Director of the MGC. Outside of work she keeps busy with her twins and their demanding softball schedules.
Vaibhav (VJ) received his M.S. in Bioinformatics in India and has been in the bioinformatics field for 13+ years. Currently, he analyzes single cell, spatial, and bulk RNA seq data. VJ likes to read, loves travelling, and finds that Indian desserts always hit the sweet spot.
Stephanie is a Lab Analyst for the Molecular Genomics Core, and the lead for 10x Genomics Single Cell RNA sequencing services provided by the core. She loves spending time with her dogs and bird watching.
Emily is a Research Lab Analyst in the Molecular Genomics Core. By day, she is the lead for spatial services within the core and by night Emily plays Dungeons and Dragons, Magic the Gathering, and watches anime.
Kevin is a bioinformatician with a focus is on the analysis of glioma data derived from many different sequencing modalities. Originally from northeast Ohio, he is an auto racing fan and once had the opportunity to drive 190mph at Daytona!
Michael graduated from Penn State (B.S., Biomedical Engineering) & Carnegie Mellon (M.S., Computational Biology). He works on single cell & spatial transcriptomic data analysis and is interested in innovative analysis methods for spatial & highly multiplexed imaging-based data.
Ellora is a Research Analyst for the Molecular Genomics Core from Calgary, Canada. She work on the MGC’s 10x Genomics single cell projects, and run other assays as needed. Ellora loves spending time outside, doing yoga, hiking, and painting.
Odmaa is a grad student from Mongolia in @DukeUPGG program (B.S. in Biology, minor in CS from NYUAD). Odmaa is interested in studying neurodegenerative disease via spatial & single cell approaches. She is passionate about STEM outreach, and enjoys painting/drawing & rock climbing
Miko, from Beijing, China, successfully completed a Ph.D. candidate in Computational Biology and Bioinformatics (CBB) at Duke. Her thesis focuses on analyzing single-cell, multiome and spatial transcriptomics data to understand brain tumor microenvironments. She’s also an avid dancer and performer!
Alan was born in Buenos Aires, then moved to NC when he was 8, which has been his home since. He is a member of the MGC, working primarily with spatial GEX platforms. When he’s not in the lab, you will find him trying not to burn down his kitchen while learning new recipes.
Julia is a joint research tech, working on various MGC projects and on the 'Risk Stratification in Children with Concussions' team for Allison Ashley-Koch's Lab. Outside of the lab she is a doting cat mom to furball Griswald and enjoys drawing, reading, yoga, and swimming.
Lauren is a graduate student in Duke University's Genetics and Genomics Program (UPGG). Originally from Jacksonville, Florida, she earned both her Bachelor’s and Master’s degrees from the University of North Florida. Her research is focused on studying the origins of glioblastoma using multi-omic approaches, aiming to deepen our understanding of this aggressive brain cancer. Outside of the lab, Lauren loves spending time in her garden, going to trivia nights, and cheering on the Jacksonville Jaguars (DTWD)!"
Emma is a grad student from South Carolina in the @DukeUPGG program and received her B.S. In Biochemistry from Clemson University. Emma is studying the lesion microenvironment in Multiple Sclerosis using single-cell and spatial transcriptomics. Outside of lab, she enjoys playing piano, drawing, and hiking.
The newest MGC member Khooshbu is a recent master’s graduate in Clinical & Experimental Therapeutics with Bioinformatics from USC. She will be working with the MGC and @prtbtcatduke on various data analysis projects. Outside of the lab, she loves painting, dancing, and cooking.














