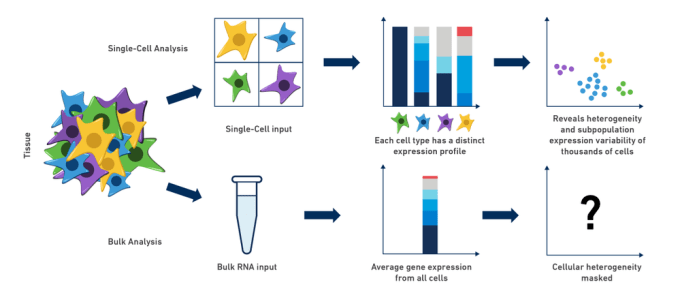
Single cell transcriptomic profiling enables characterization of normative gene expression and stratification of complex cell types from heterogeneous tissue, identifying rare cell types & characterizing subpopulations of seemingly homogeneous cell populations.
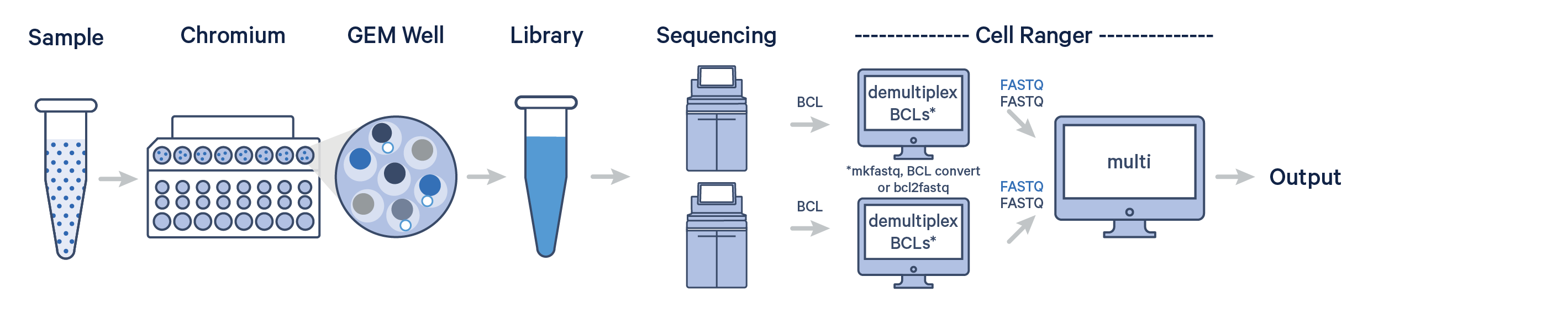
The 10x Genomics Chromium X platform partitions between 500-20,000 single cells/nuclei into Gel Beads-in-emulsion (GEMs), using a microfluidics-based instrument & nanoliter-scale gel bead technology. The GEMs are then processed through reverse transcription (GEM-RT) and cDNA amplification before proceeding to library preparation. A common 10x cell Barcode is introduced during GEM generation, allowing tracing of individual reads back to each cell during analysis.
10x Genomics’ 3’ scRNAseq assays offer scalable profiling of 3’ gene expression using ~3.6M separately sampled 10x Barcodes to index each cell’s transcriptome. The MGC has been running scRNAseq since 2016, ranging across many species and cell types, to generate over 2000 single cell libraries.
- Input: fresh single-cell suspension (>80% viable) or cryopreserved single-cell suspension
- Add ons - Feature Barcoding for cell surface proteins or CRISPR screening on customer provided sgRNA positive cells
- Customer is responsible for selecting & ordering Biolegend Totalseq antibodies, antibody titration, and staining prior to delivery of samples for scRNAseq run
- If using FACs, note that we typically see ~50% overestimation of cell counts off the sorter
- We offer free cell assessments (count and viability) prior to your scheduled project
10x Genomics’ 3’ snRNAseq assays enable flexibility when challenges with fresh biospecimen arise due to storage conditions, or tissue dissociation that may cause lysing of some cell types. snRNAseq samples require nuclei isolation optimization to maximize input sample quality: prior to run date, either the MGC will run nuclei isolation optimization tests, or customer will bring samples for counting & nuclei intactness assessment.
- Input - cells, frozen tissues, or organoids for nuclei isolation
- For frozen tissue, we recommend RNA extraction and QC, with a RIN >=8
- Validated 10x nuclei isolation protocols for wide variety of sample types
- Lysis optimization includes testing of lysis times, buffer concentrations, centrifugation speeds, filtrations, and wash steps to produce high quality nuclei samples (4 samples/optimization)
- A quality nuclei suspension should have <5% viable cells with >80% of the nuclei intact
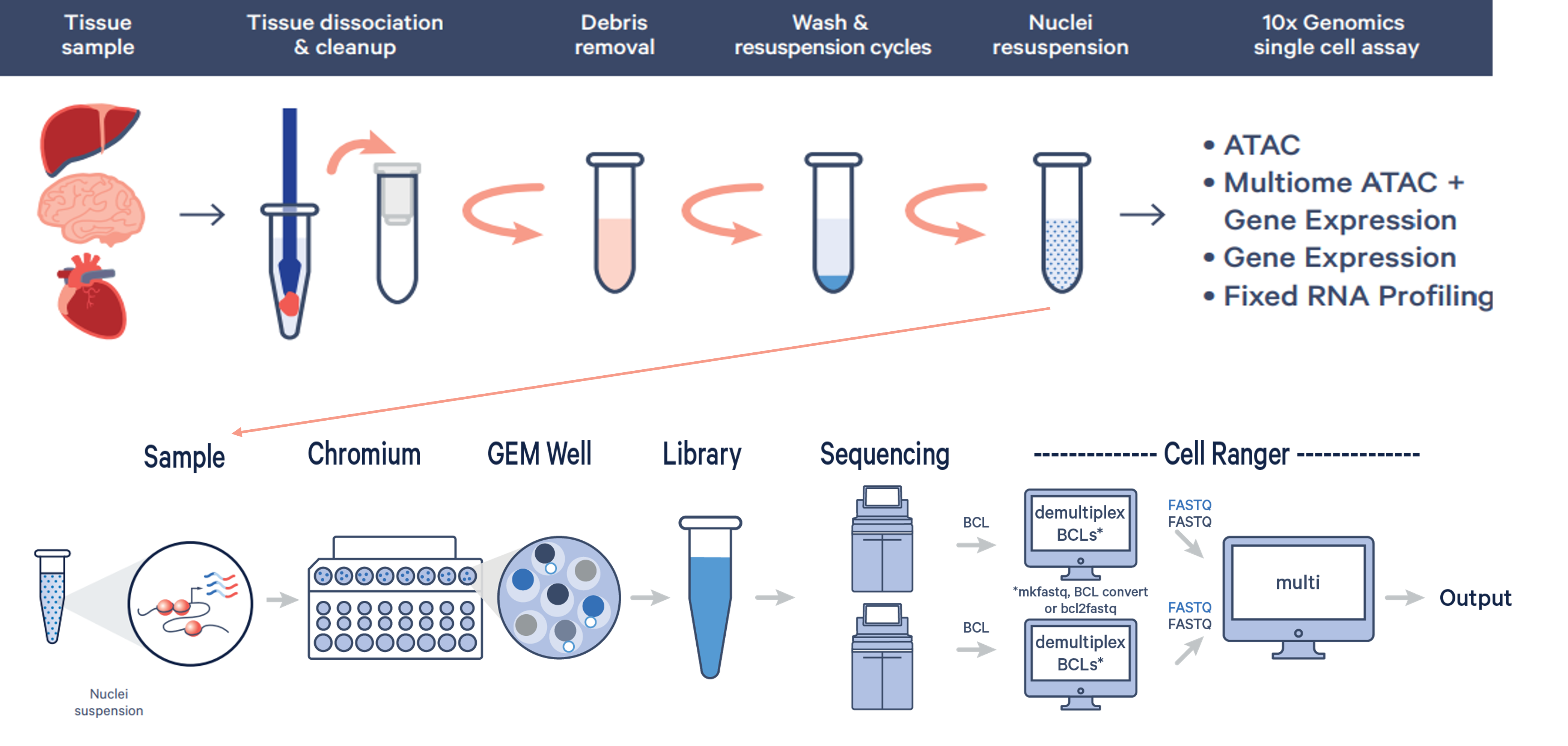
Single Nuclei Workflow (Images via 10x Genomics)
The 10x Genomics scRNAseq Flex assay provides flexibility during sample preparation and experimental design by enabling batching across time and collection sites, cell surface protein labelling, sample multiplexing with unique sample barcodes, infectious agent neutralization, and the potential for cost savings. Gene expression in the Flex assay is measured using probe pairs, designed to hybridize to mRNA specifically. During GEM generation, the probes are ligated and the 10x Barcode is added. The barcoded product is then amplified to produce gene expression (GEX) libraries.
- For more information on Flex probe sets, click here
- Validated for cells & nuclei fixed with formaldehyde or isolated from FFPE tissue blocks
- Enables investigators to profile archival banked FFPE samples
- Mostly nuclei will be isolated from FFPE tissue blocks (mix of cells & nuclei)
- We recommend RNA extraction and QC, with a DV200 >=30%
- Requires less sequencing depth than traditional scRNAseq, meaning lower sequencing costs
- Covers 18,000 human genes or 19,000 mouse genes
- Target 500 - 128k cells per chip with multiplexing and pooling capabilities (up to 10k/sample)
- Input – cells, nuclei or tissues fixed with 10x Genomics’ Chromium Next GEM Single Cell Fixed RNA Sample Preparation Kit, or FFPE scrolls
- Samples can be fixed & stored for 6 months at -80°C
- FFPE scrolls cut for dissociation can be stored for 1 year at 4°C
- Feature barcoding available to measure protein expression via oligo-conjugated nucleotides
- Barcode oligo capture: prior to GEM generation, labelled & fixed suspensions are hybridized with Probe Barcodes (& Antibody Multiplexing Barcodes, if needed), which are then amplified to produce protein expression libraries alongside GEX libraries
- TotalSeq-C compatible for single & multiplex assay
- TotalSeq-B only compatible with singleplex assay
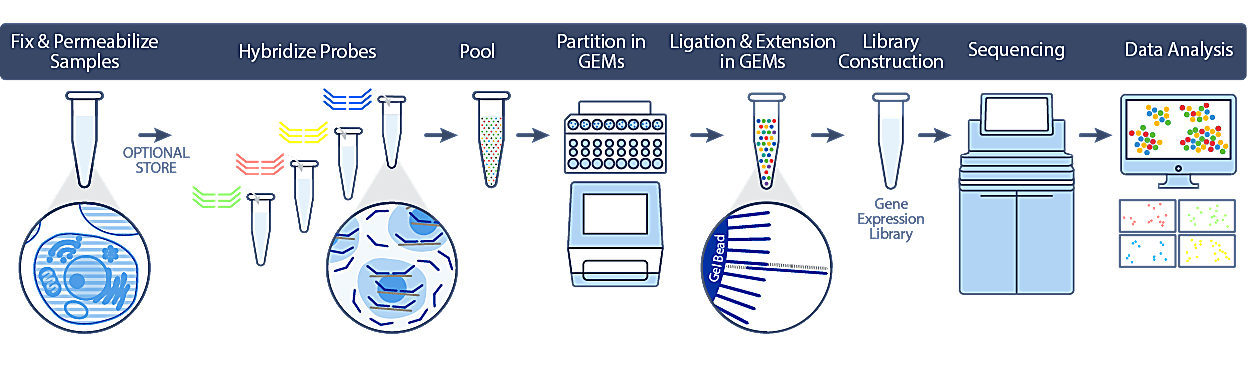
Flex workflow (Image via 10x Genomics)
The 10x Genomics Single Cell V(D)J assay profiles 500-20,000 individual cells per sample with paired T-cell receptor (TCR) and/or B-cell immunoglobulin (Ig) receptor sequences, while other immune profiling approaches are often limited to heavy chain profiling of specificity conferring regions of the genome.
Unlike other 3’ 10x Genomics scRNAseq assays, the 5’ protocols profile full-length transcripts (5' UTR to constant region).
Following GEM-RT and cDNA amplification, V(D)J enrichment is performed on full-length cDNA using primers specific to TCR and/or BCR constant regions, facilitating the generation of both V(D)J & 5’ GEX libraries. If both T and B cells are expected in a cell population, TCR and Ig transcripts can be amplified in separate reactions from the same amplified cDNA material to yield distinctive TCR & BCR libraries.
- Input – fresh single-cell suspension (>80% viable) or cryopreserved single-cell suspension
- Add ons - Feature Barcoding for cell surface proteins or CRISPR screening
- Customer responsible for selecting & ordering Biolegend Totalseq antibodies, antibody titration, and staining prior to delivery of samples for scRNAseq run
- If using FACs, note that we typically see ~50% overestimation of cell counts off the sorter
- We offer free cell assessments (count and viability) prior to your scheduled project
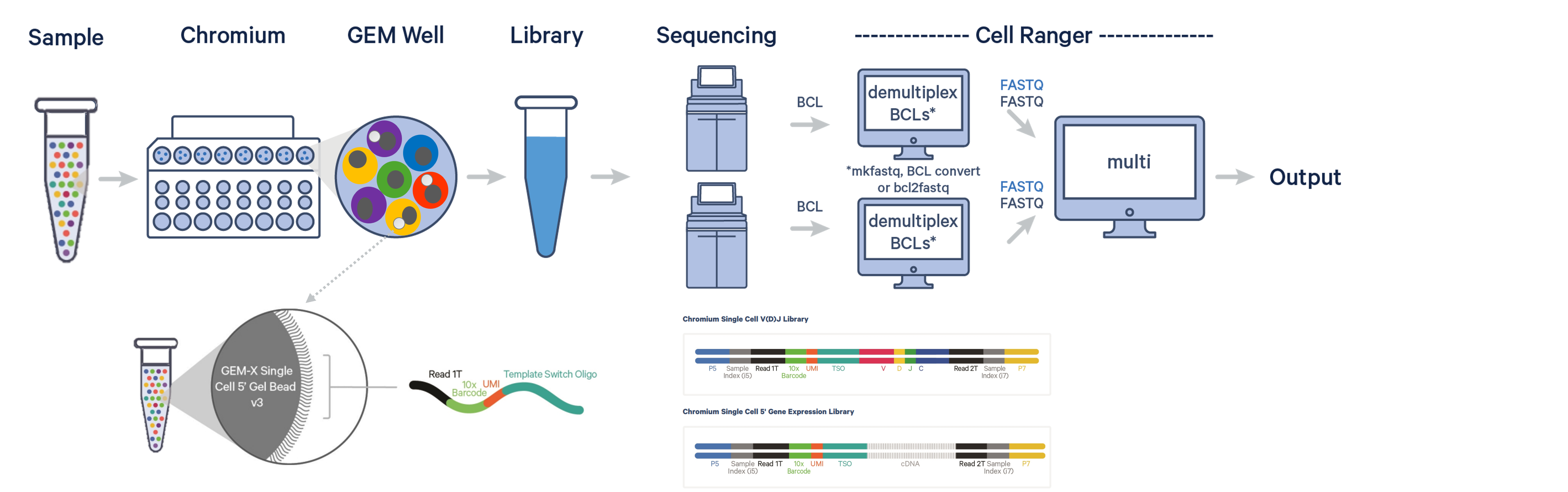
5' Single Cell Workflow
Chromatin alterations within the genome can reveal a great deal about the state of the cell. With as little as a few hundred cells, 10x Genomics scATAC-seq recognizes accessible chromatin regions and identifies rare and important cellular subtypes, providing an unprecedented view of heterogeneous chromatin regulation. After counting and quality assessment, samples undergo a transposition step prior to GEM-RT, preferentially fragmenting DNA in open chromatin regions and adding adapter sequences to the ends of the DNA fragments.
- Input - cells, frozen tissues, or organoids for nuclei isolation
- Refer to snRNAseq section for more information on nuclei isolation optimization
- ATAC protocol available to be run individually or as Multiome run (see below)
snRNAseq Multiome (ATAC & Gene Expression):
The Chromium single cell Multiome ATAC + Gene Expression assay allows investigators to leverage two modalities at once, uniting transcriptional and chromatin landscapes from a single nucleus to produce both GEX & ATAC libraries. Characterization of direct linkages between gene expression and regulatory elements provides an in-depth view of the cell state, differential expression drivers and functional differences within cell populations.
- Input - cells, frozen tissues, or organoids for nuclei isolation
- Refer to snRNAseq section for more information on nuclei isolation optimization
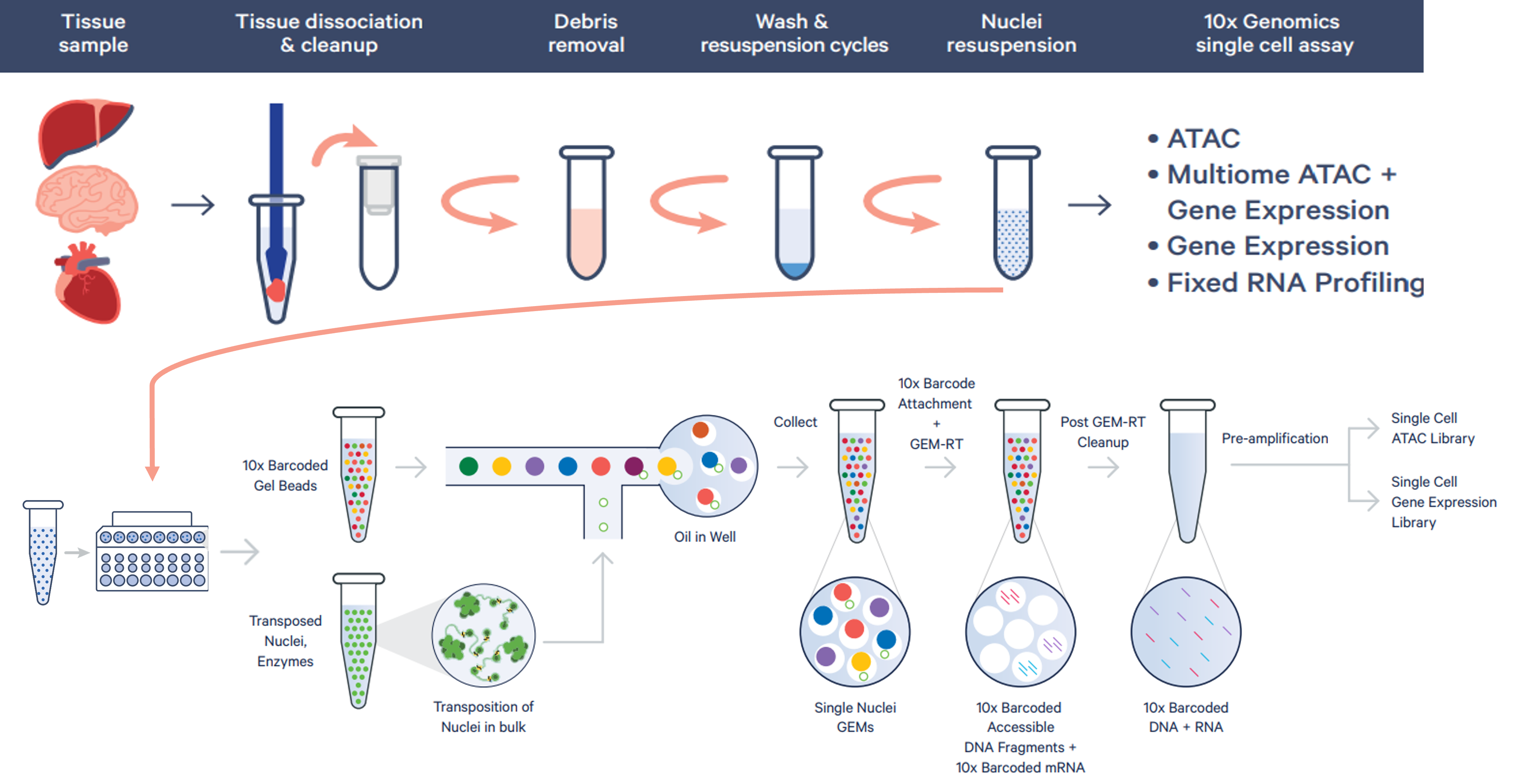
Multiome Workflow (Images via 10x Genomics)
- Refer to snRNAseq section for more information on nuclei isolation optimization
To ensure the highest quality of service and successfully meet the needs of your project, we require specific information from you. Without your completed sample submission forms and service requests, we are unable to proceed. Please let us know if we can assist you in any way.
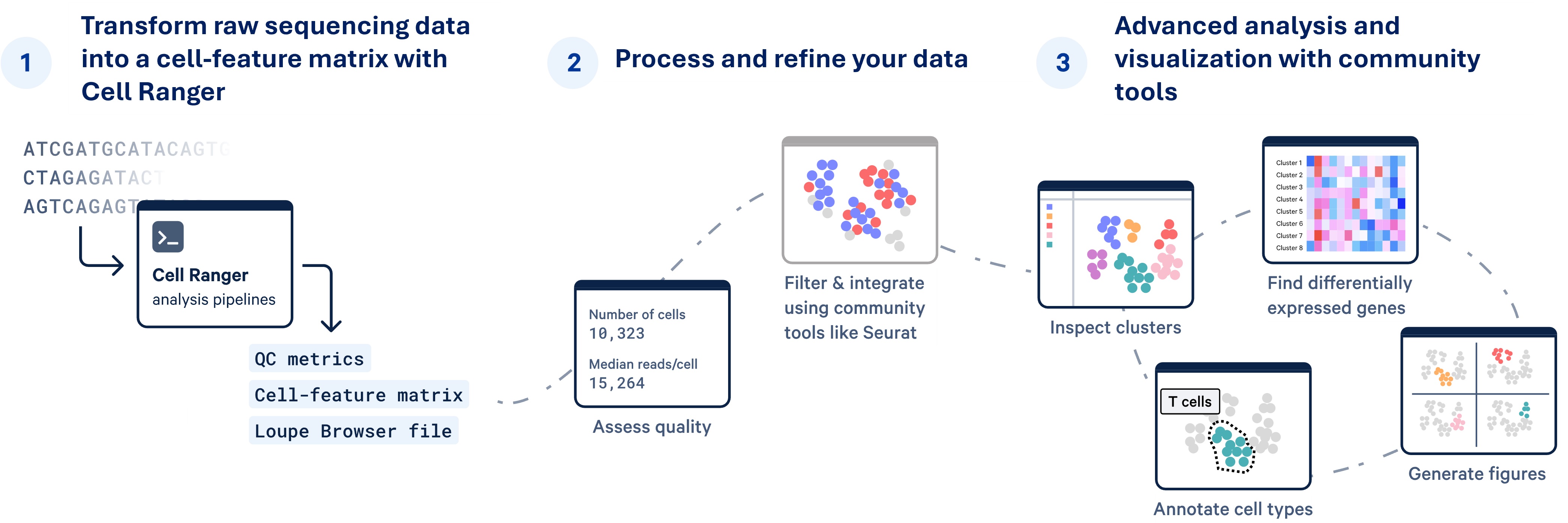
Basic Analysis (10x Genomics Cell Ranger) - required
FastQC: conversion of raw sequencing data to Fastq format & alignment of sequencing reads to reference transcriptome
- Raw data will be uploaded to SFTP server, as a bcl file - login information to access data will be sent by email to MGC bioinformaticians & customer to download (raw data stored for 30 days on SFTP server)
CellRanger: run a set of analysis pipelines, processing scRNA-seq data to align reads, generate feature-barcode matrices, and perform clustering and gene expression analysis
- CellRanger web summary metrics are assessed to determine the success of sequencing, mapping, and cell metrics
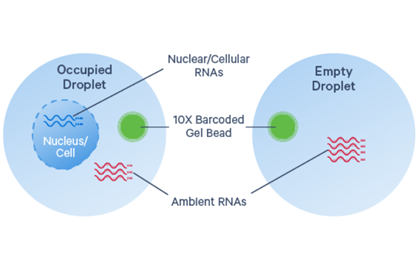
CellBender Filtering (if required) - deep generative model distinguishes cell-containing droplets from empty ones, learns the profile of background noise, and retrieves cleaner gene expression profiles by removing ambient RNA
- Ambient RNA: free-floating transcripts from cell-free RNA molecules, ruptured/dead/dying cells in suspension, or debris, captured in GEMs and resulting in low level background RNA counts
- High background counts tend to occur more frequently in certain tissue types, particularly when working with single cell nuclei samples, due to cytoplasmic RNA in solution from cell lysis
Deliverable files include the following:
- Web summary files
- Count matrix files
- Alignment files
- FASTQ files
Advanced Analysis (SoupX, Seurat, SingleR, scType, Doublet finder) - if requested
- Cell count correction using SoupX
- Quality control filtering
- Dimensional reduction and cell clustering
- Doublet removal using doublet finder
- Conserved gene marker identification per cell cluster
- Differentially expressed gene identification between samples
- Cluster annotation using automatic prediction algorithm (singleR/sctype)
- Plotting initial genes of interest (up to 30)
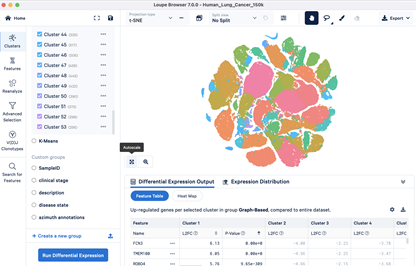
Deliverable files include the following (as requested):
- Clustering UMAP plots
- QC plots
- Cell count info in CSV and bar graph
- Heatmap of top 10 genes per cluster
- Differential gene expression results per cluster in CSV and between samples if comparing two samples
- Average gene expression per cluster in CSV
- Gene's expression in feature and violin plots
Other Analyses - if requested

- Gene Set Enrichment Analysis (GSEA) between the identified clusters or between the samples
- RNA Velocity analysis using Velocyto
- Pseudotime analysis using Monocle
- Cell interaction and signaling network analysis using Cellchat
- Gene ontology enrichment analysis using Enrichr/Clusterprofiler
- Pseudobulk differential gene expression analysis using Muscat
- Gene regulatory network analysis using WGCNA/SCENIC
- Copy number variation analysis using InferCNV
- Figures for papers/grants that aren’t part of the initial analysis