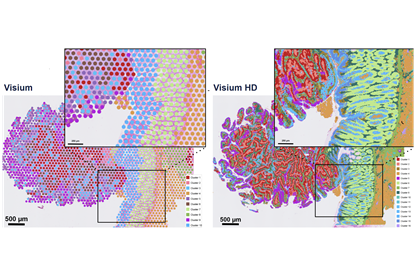
A variety of spatial-omics assays facilitate characterization of cell-cell and cell neighborhoods during critical phases of development and disease. The MGC offers two spatial profiling solutions that can assay RNA at or near single cell resolution.
Xenium Human GBM Image Courtesy of Simon Gregory
In Situ Sequencing
Released in 2023, the 10x Genomics Xenium Platform is a leading-edge spatial transcriptomics technology that allows investigators to visualize target transcripts at subcellular resolution within a spatial context. The Xenium Analyzer detects each fluorescently labeled probe captured in microscopic images of the tissue of interest in a cyclical imaging scheme to create a unique optical signature for each target. For a full list of available panels, click here.
- Validated for fresh frozen or FFPE tissue sections
- ROI must fit within 10.45x22.45mm capture area
- Can fit multiple smaller sections of tissue into one capture area
- Facilitates visualization of up to 5000 genes/run
- Focused on targeted, customizable panels
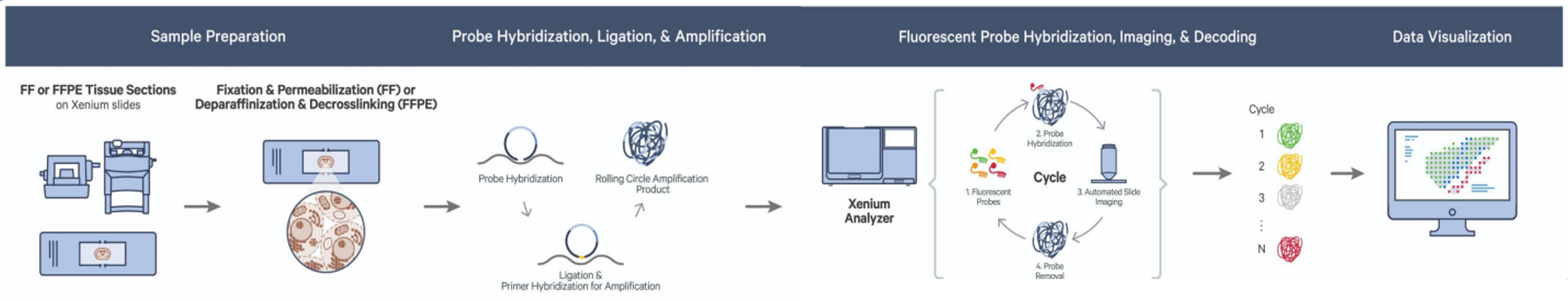

Alongside the spatial omics services listed in the sidebar, here are the Xenium-specific services offered by the MGC:
- Tissue block DAPI QC
- Multimodal Cell Segmentation staining
- Xenium Analyzer Imaging and output
- Post-Xenium H&E staining
Spatial Profiling of RNA

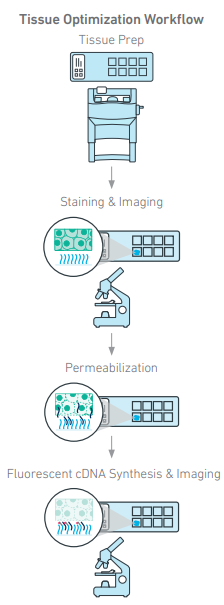
The 10x Genomics Visium platform provides morphological context of cellular function in tissue microenvironments by incorporating total mRNA analysis of intact tissue sections. 10x Genomics has developed multiple workflows to provide flexibility for sample preparation and experimental design. Each Visium workflow follows a standard histological process: sectioning, tissue preparation, staining (H&E or IF), and imaging, followed by Visium analyte capture and processing into NGS library preparation.
- Species-agnostic
- Utilizes 10x Genomics Gene Expression Slides with four 6.5mm2 fields per slide
- Compatible with Fresh-frozen tissue
- Resolution of 50um spots, 100um center to center distance
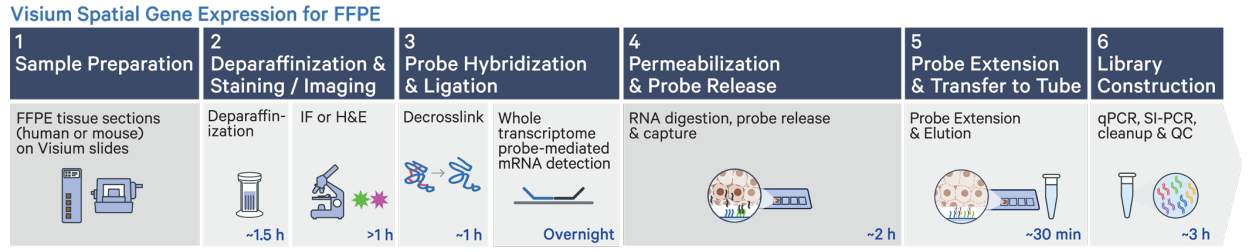
- Validated for human & mouse FFPE, Fresh Frozen and Fixed Frozen tissue
- Designed to accommodate fragmented RNA
- Enables investigators to profile archival banked FFPE samples
- Uses a panel of 25mer probes to target 18-20k genes across the transcriptome
- Resolution of 50um spots, 100um center to center distance
- Customized probe spike-in option available
- Utilizes regular glass slides for sample preparation to transfer analytes to either 6.5mm2 or 11mm2 gene expression slide fields.
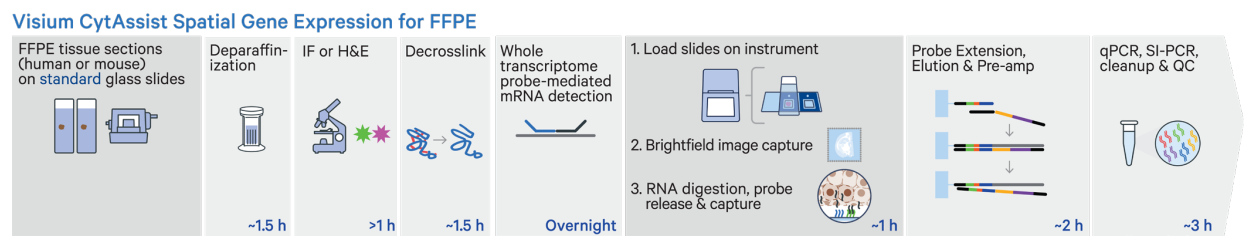
- Validated for human & mouse FFPE, Fresh Frozen and Fixed Frozen tissue
- Designed to accommodate fragmented RNA
- Enables investigators to profile archival banked FFPE samples
- Uses a panel of 25mer probes to target 18-20k genes across the transcriptome
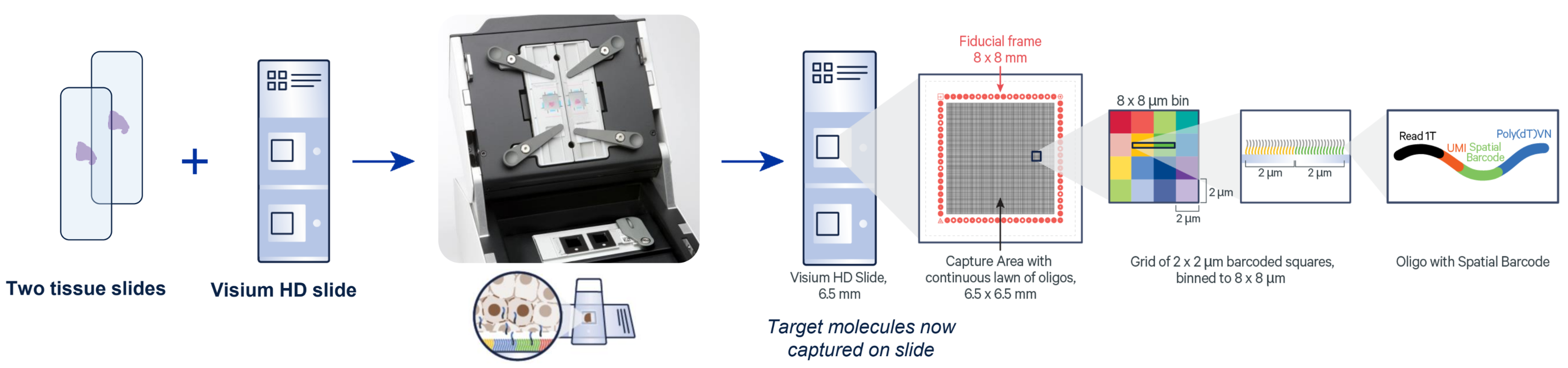
- Resolution of 2um squares, no space between (full coverage)
- Customized probe spike-in option available
- Utilizes regular glass slides for sample preparation to transfer analytes to 6.5mm2 gene expression slide fields.
Alongside the spatial omics services listed in the sidebar, here are the Visium-specific services offered by the MGC:
- Tissue Optimization Assay (Visium Direct Placement, required)
- H&E staining
- Next Gen Sequencing (outsourced)
- Basic data analysis (Spaceranger)
To ensure the highest quality of service and successfully meet the needs of your project, we require specific information from you. Without your completed sample submission forms and service requests, we are unable to proceed. Please let us know if we can assist you in any way.
Visium Basic Analysis (10x Genomics Space Ranger) – required
FastQC: conversion of raw sequencing data to Fastq format, alignment of sequencing reads to reference transcriptome, and tissue/fiducial alignment to input slide
- Raw data will be uploaded to SFTP server, as a bcl file - login information to access data will be sent by email to MGC bioinformaticians & customer to download (raw data stored for 30 days on SFTP server)
Space Ranger: generation of count matrix and alignment files for each sample
Space Ranger web summary metrics are assessed to determine the success of sequencing, mapping, and alignment.
Deliverable files include the following:
- Count matrix files
- Alignment files
- Loupe Browser slide image
- Fastq files (if required)
Visium Advanced Analysis (Banksy, SpaceXR, C-Side) - if requested
- Quality control filtering
- Basic filtering
- Non-HD (Seurat): select thresholds and filter based on number of counts per UMI, number of unique genes, and mitochondrial percentage
- HD (Enact software): converts bins to cells, performs cell annotation and filtering
- Basic filtering
- Dimensional reduction and cell clustering with Banksy
- Conserved gene marker identification per cell cluster
- Differentially expressed gene identification between samples by cluster
- Plotting initial genes of interest (up to 30)
- Deconvolution using relevant single-cell reference with SpaceXR
- Differentially expressed genes by deconvoluted cell-type with C-Side (depends on amount of celltype present whether or not it can be performed)
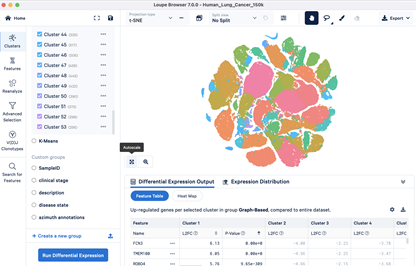
Deliverable files include the following:
- Results from Seurat, Banksy, and SpaceXR analyses (.csv, .png, .pdf, .rds files)
Other Visium Analyses – if requested
- Gene Set Enrichment Analysis (GSEA) between the identified clusters or between the samples
- Cell-cell communication with COMMOT
- Spatially variable gene expression (per slide basis) with SpaceXR
- Spatial Trajectory Analysis with spaTrack
Xenium Basic Analysis (10x Genomics Xenium Explorer) – required
Xenium Explorer: imaging, transcript decoding, cell segmentation, and cell transcript assignment
Verify transcript counts and cell segmentation via web summary files
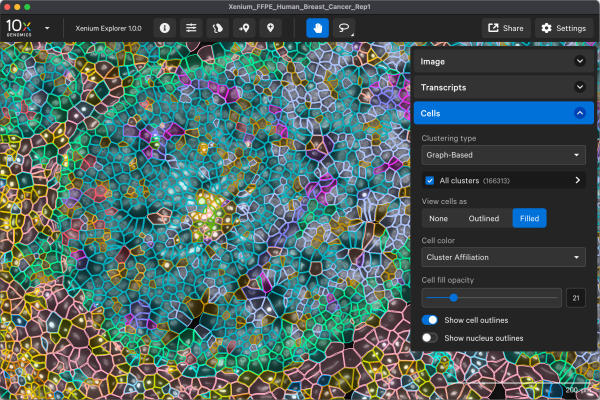
Deliverable files include the following:
- Image files
- Matrix files
- Cell and transcript info files
Xenium Advanced Analysis (SpaceXR Seurat Wrapper) - if requested
- QC analysis – filtering outliers (Basic filtering: spots <10 counts, <5 expressed genes)
- Normalization
- Unsupervised clustering
- Gene expression plots (All genes)
- Using reference single cell data - SpaceXR Seurat wrapper
- Comparative analysis between regions/samples to find DEG.
- GSEA and volcano plots for DEGs
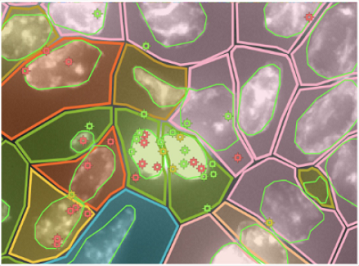
Deliverable files include the following:
- Clustering UMAP/Spatial plots
- QC plots
- Cell count info in csv and bar graph
- Heatmap of top 10 genes per cluster
- Differential gene expression results per cluster in csv and between regions of interest (ROI) if comparing two ROIs
- Average gene expression per cluster in csv
- Genes expression in feature
- Violin and spatial plots for all genes in the panel
- UMAP/spatial plots from cell type prediction
- Volcano and pathway plots from DEG results
Other Xenium Analyses – if requested
- Neighborhood enrichment analysis (Squidpy)
- Co-occurrence probability analysis (Squidpy)
- Gene spatial correlation analysis (Squidpy)
- Cell- cell interaction analysis (stLearn)
- Spatial trajectory inference (spaTrack)
- Tissue Domain Identification (Bansky)